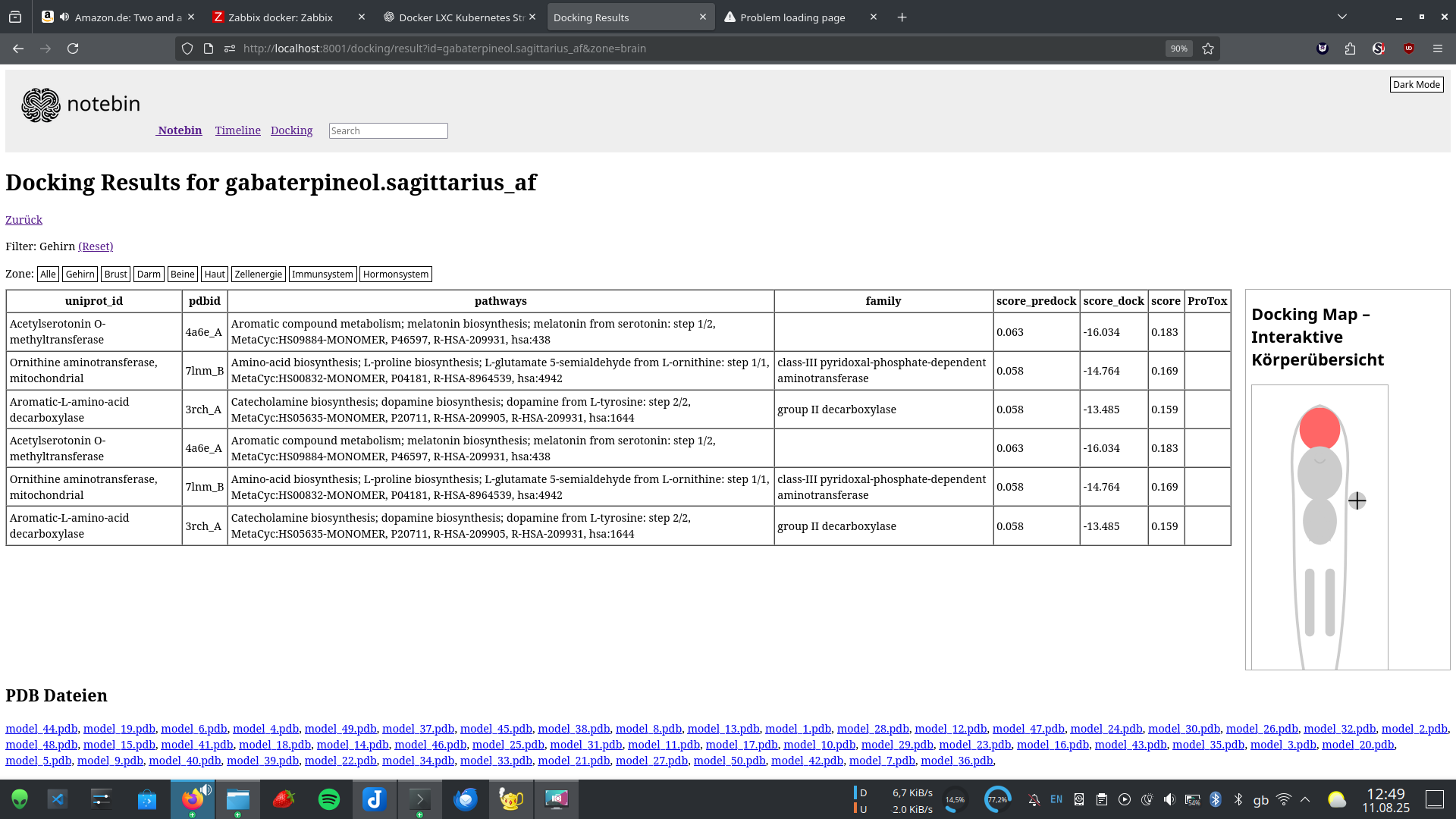
Dockbin - Dynamic Omics & Knowledge-Based Immune Network
Von Mikroplastik bis Medikament – wir kartieren, wie Substanzen mit dem menschlichen Proteom interagieren.
Why I’m Doing This
I’m living with CFS myself, which gives me a strong, personal motivation to develop Dockbin AI-driven Exposomics & Proteomics. Beyond my own condition, I want to better understand the complex interactions within the human body—how molecular systems, immune networks, and environmental factors influence each other. This platform is designed to visualize and analyze these interactions, supporting research that can ultimately lead to new insights into systemic biology and potential therapeutic approaches. By integrating AI-driven analysis with comprehensive proteomics and exposomics data, Dockbin aims to make these relationships accessible for scientific study, helping researchers uncover patterns that are otherwise difficult to detect. My personal experience with CFS drives this work, ensuring that it is both practical and oriented toward real-world biomedical challenges.
Kernidee
Unsere Plattform kombiniert eine Datenbank von Substanzen mit KI-gestütztem Docking und Visualisierung. Ein Alleinstellungsmerkmal ist die Integration synthetischer Exposome-Bibliotheken (z. B. randomisierte Mikroplastik-Fragmente), die mit z.B. Olink® oder anderen natuerlichen Proteomdaten verknüpft werden.
Methodik
- KI-Layer erzeugt Embeddings, Cluster & Docking-Vorhersagen
- Substanzen: therapeutische Kandidaten + synthetische Umweltstressoren
- Visuelle Interaktionskarten für Forscher:innen
Impact
Unser Ansatz erweitert Proteomics um den Bereich Exposomics: Wir zeigen, wie Umweltfaktoren (z. B. Mikroplastik) und Medikamente gleichermaßen in Proteom-Netzwerke integriert werden können.
So entstehen neue Hypothesen zu Immunregulation, Neuroimmun-Crosstalk und Wirkstoffentwicklung.
Kontakt & Kollaboration
Wir stehen für Open-Science-Kooperationen bereit – von Forschungsgruppen bis Förderinstitutionen.
Background & Rationale
Proteomics has become an indispensable tool for understanding the molecular underpinnings of health and disease. However, the majority of computational pipelines remain narrowly focused on either clinical biomarkers or drug target discovery. Environmental health – and especially the integration of emerging exposome stressors such as microplastic derivatives – is still largely absent from proteomic modeling frameworks.
This represents a significant gap: immune and neuro-immune regulation are not only influenced by classical disease pathways, but also by chronic, low-level exposure to environmental perturbants. Bridging this gap requires tools that are both flexible and interdisciplinary, connecting biomedical proteomics with toxicology, ecology, and drug discovery.
Project Vision
We are developing a platform that combines structured substance databases, AI-driven docking pipelines, and interactive visualization to generate proteome-level interaction maps. A distinguishing feature of our approach is the ability to generate synthetic exposome libraries, such as randomized microplastic fragments or derived oligomers, and integrate them alongside pharmacological compounds.
This dual capability allows us to explore:
-
Pharmacological hypotheses (drug–protein docking, network connectivity, biomarker validation).
-
Environmental impact hypotheses (immune and neuroimmune responses to synthetic particles and environmental derivatives).
By demonstrating feasibility across both domains, our platform can significantly expand the utility of proteomic data for translational research.
Technical Approach
Our running prototype already supports:
-
Data import and preprocessing – scalable pipelines for proteomic datasets (Olink® Explore HT as target input).
-
Saggitarius_AF from seoklabs, as Proteom Docking engine
-
AI-embedding layer – transformation of high-dimensional protein expression profiles into machine-learning–ready vectors, enabling clustering, anomaly detection, and predictive docking.
-
Docking engine integration – flexible docking that supports both therapeutic molecules and synthetic exposome constructs.
-
Visualization layer – interactive mapping of protein–compound interaction networks, designed to be accessible for both domain specialists and interdisciplinary users.
Planned Data Integration
We have entered the Olink® competition to access Explore HT data (5,400 proteins across 86 serum samples). The dataset is ideally suited for this proof-of-concept study: sufficiently broad to capture immunological and neuroimmunological signatures, yet compact enough for rapid prototyping and technical validation.
Pending approval, these data will serve as the empirical backbone for both of our project strands:
-
Neuromorphic, photonically-driven processor models (requiring realistic protein-level dynamics as input for adaptive regulatory mechanisms).
-
Proteomic docking and exposome mapping (testing novel environmental and pharmacological compounds against immune system signatures).
Impact
The impact of this project is dual and synergistic:
-
For biomedical research: We create a new class of intuitive proteomic interaction maps that can accelerate hypothesis generation and drug discovery.
-
For environmental health: We pioneer the integration of exposome stressors into proteomic models, extending proteomics beyond clinical and pharmaceutical contexts.
By connecting these two domains through a common data foundation, we catalyze new insights into immune regulation, neuro-immune cross-talk, and the effects of novel environmental exposures.
Dissemination & IP Strategy
-
Software, pipelines, and visualization tools will be openly accessible (Open Science principle).
-
Hardware architectures (neuromorphic processor models) will be protected under proprietary IP.
-
Dual-use of Olink® datasets will be explicitly communicated as an efficiency and innovation strategy.
Status
-
dead until further notice. health issues and collabs failed :(
meh.